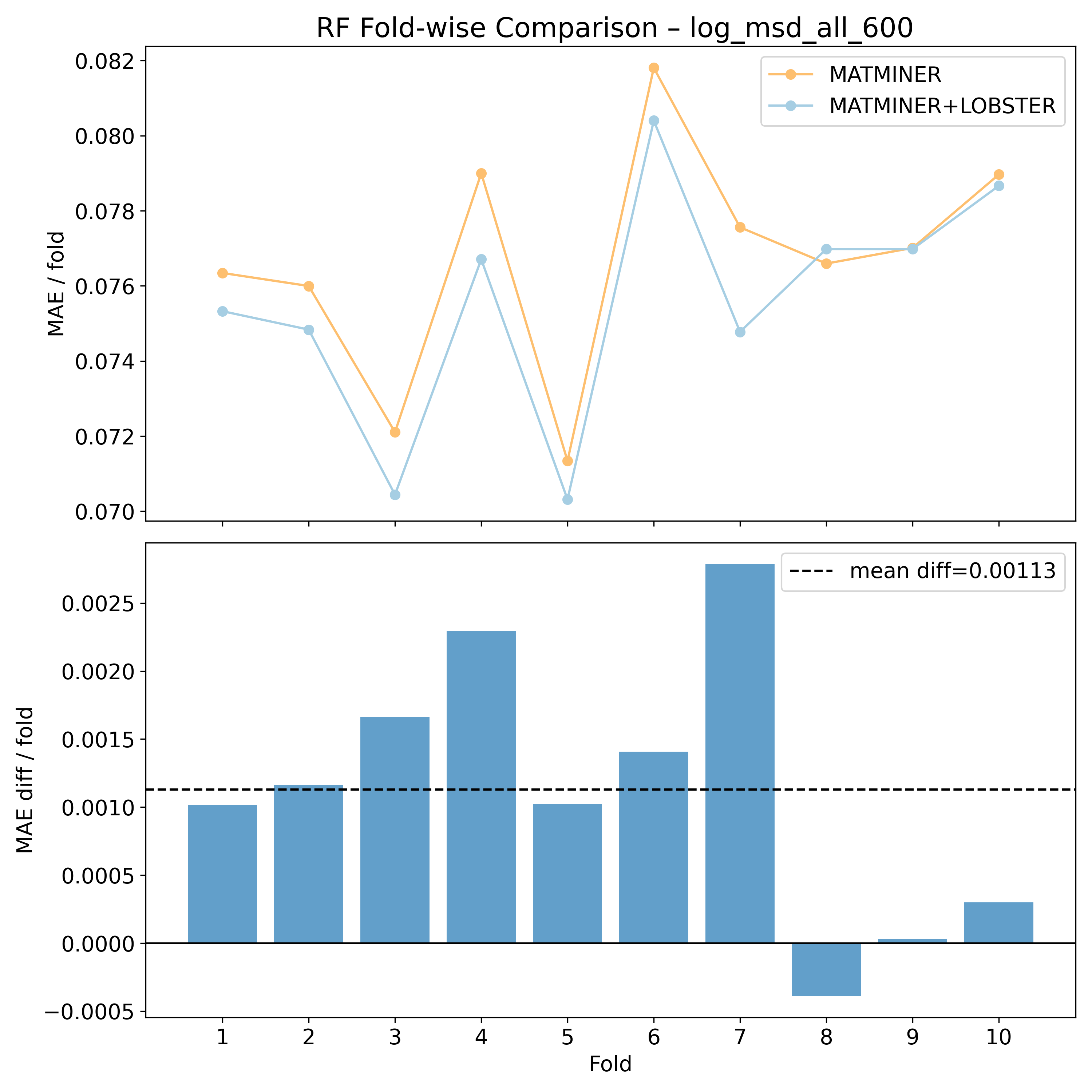
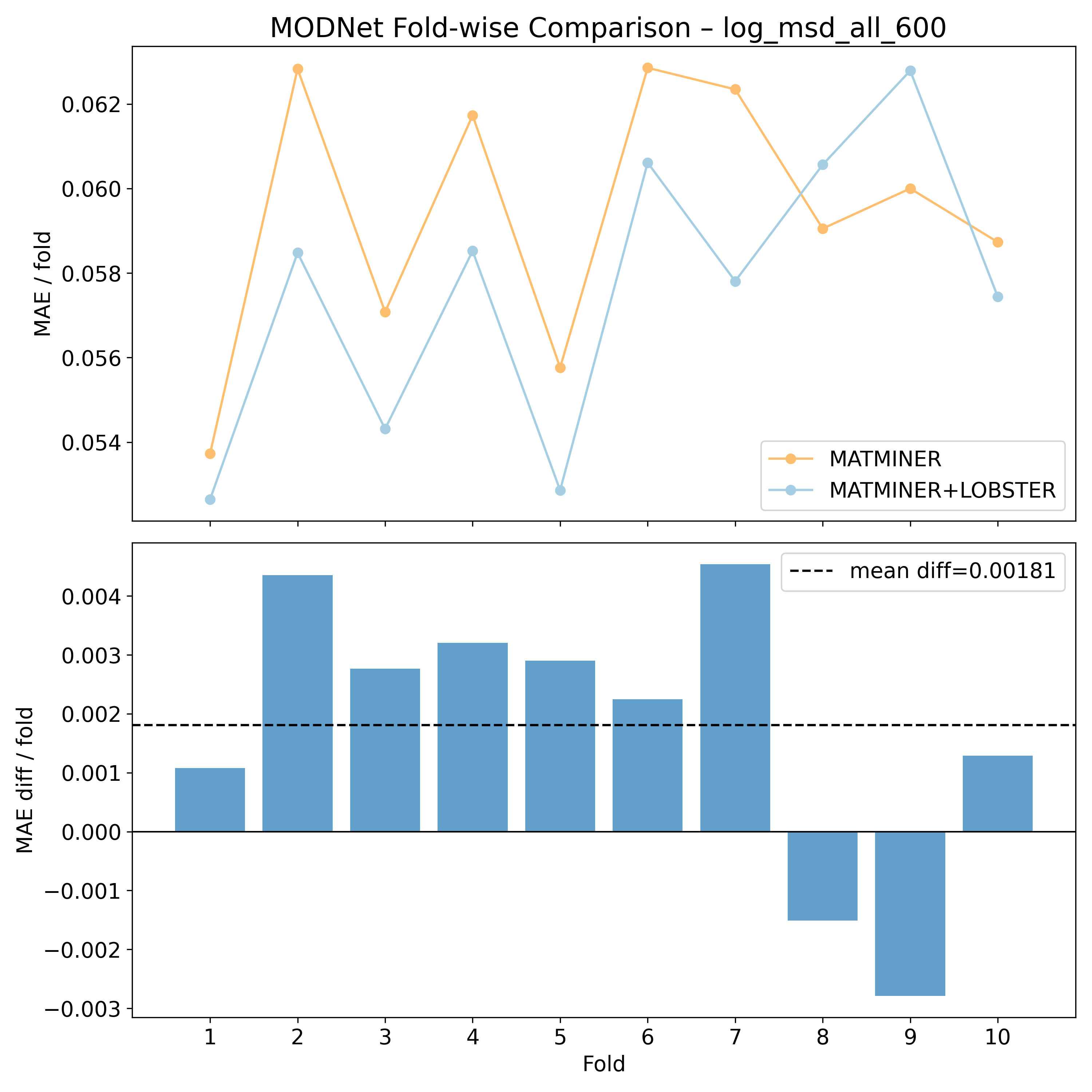
Log10 (All unique sites mean squared displacements @ 600K - Ų) - log_msd_all_600#
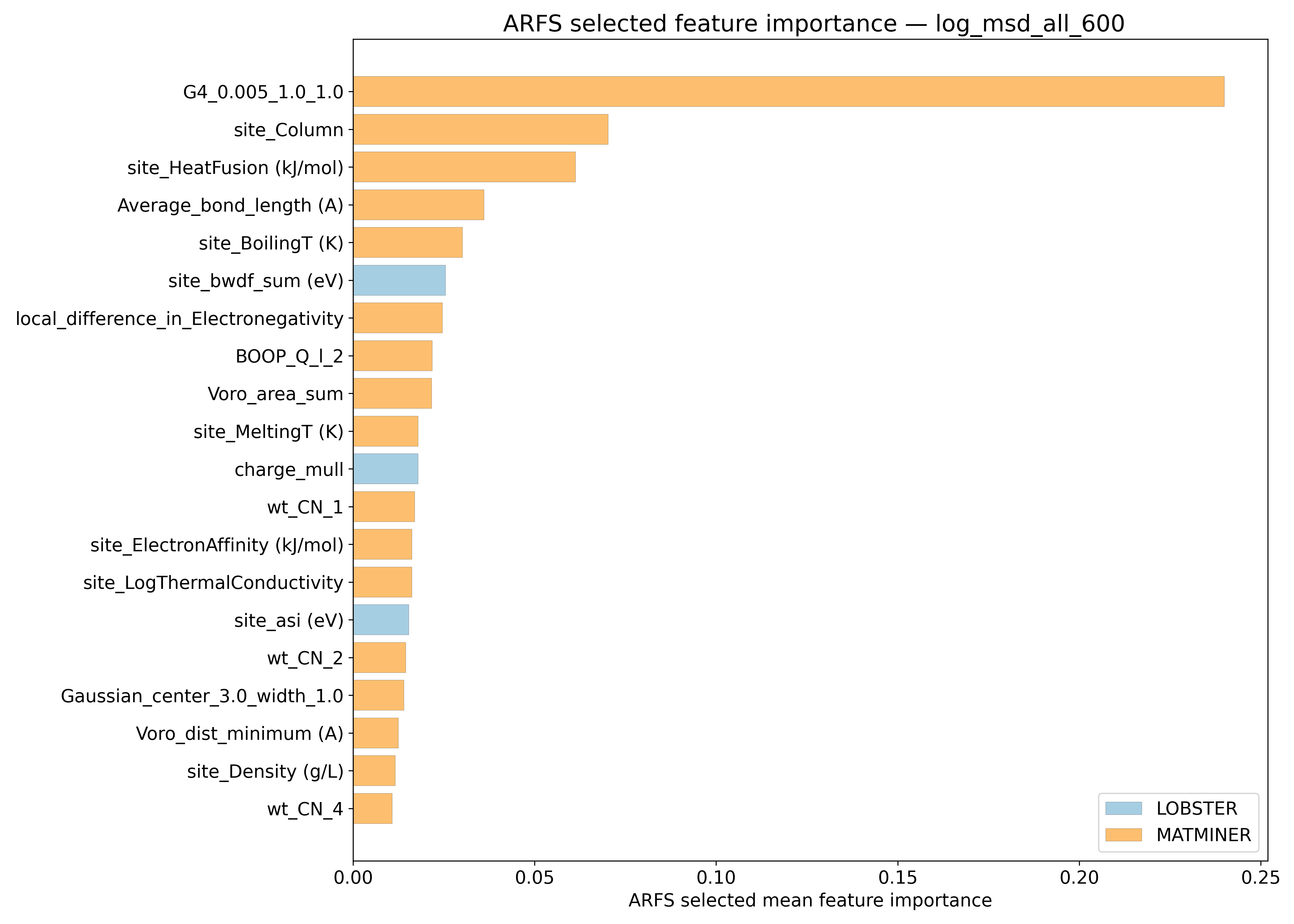
ARFS Top features#
ARFS selected descriptors#
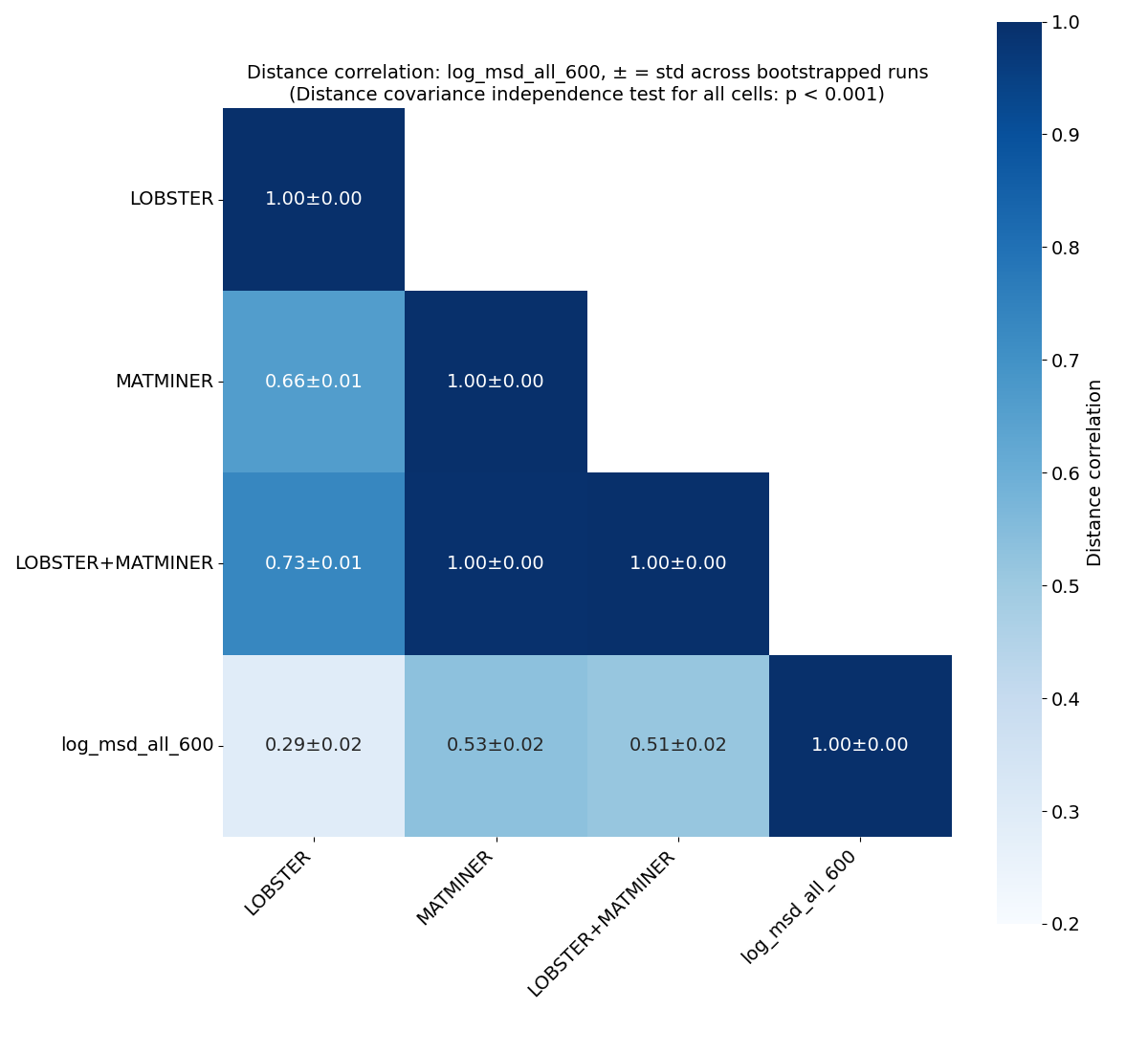
Correlation analysis#
Distance correlation#
Dependency graphs#
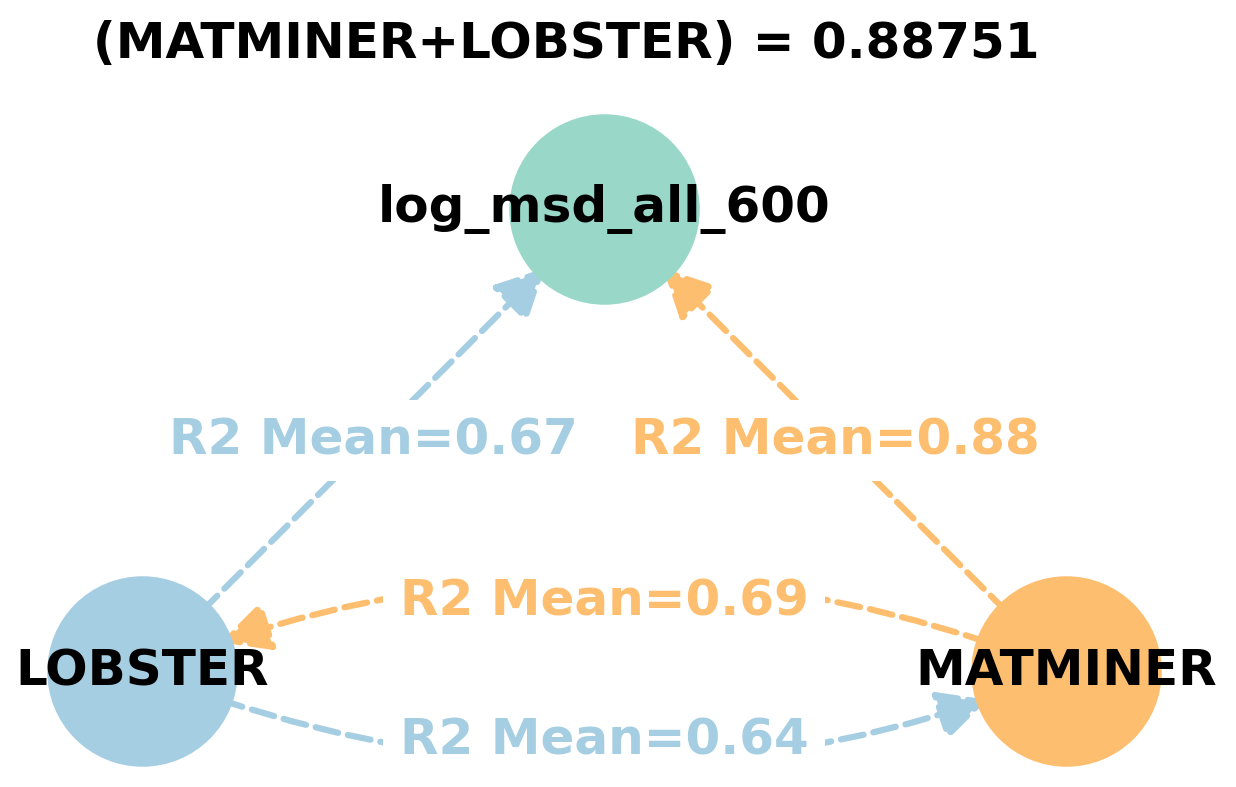
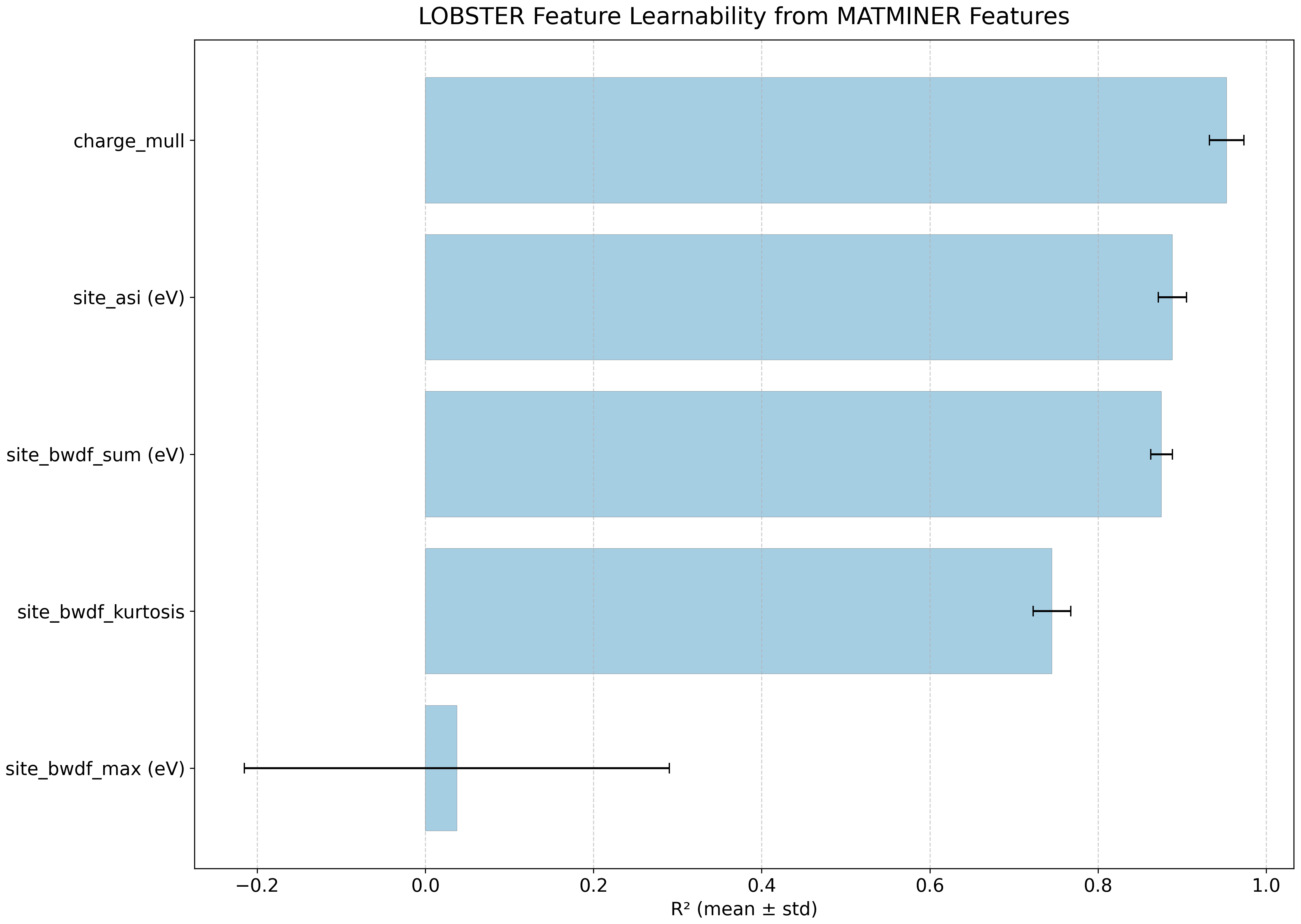
Feature learnability#
Model performance#
5-Fold CV Metrics overview#
RF - MATMINER
train_rmse |
test_rmse |
train_errors |
test_errors |
train_r2 |
test_r2 |
|
|---|---|---|---|---|---|---|
mean |
0.04056 |
0.10898 |
0.02936 |
0.07886 |
0.98396 |
0.88382 |
min |
0.04 |
0.1048 |
0.0291 |
0.078 |
0.9834 |
0.8696 |
max |
0.0411 |
0.1143 |
0.0296 |
0.0803 |
0.9845 |
0.895 |
std |
0.000397995 |
0.00360411 |
0.000215407 |
0.000776144 |
0.000361109 |
0.00866912 |
RF - MATMINER+LOBSTER
train_rmse |
test_rmse |
train_errors |
test_errors |
train_r2 |
test_r2 |
|
|---|---|---|---|---|---|---|
mean |
0.03982 |
0.10732 |
0.02878 |
0.07774 |
0.98452 |
0.88724 |
min |
0.0393 |
0.1018 |
0.0286 |
0.0761 |
0.9842 |
0.8724 |
max |
0.0401 |
0.113 |
0.0291 |
0.0801 |
0.985 |
0.8989 |
std |
0.000292575 |
0.00462662 |
0.000193907 |
0.00145959 |
0.000263818 |
0.00959679 |
MODNet - MATMINER
train_rmse |
test_rmse |
train_errors |
test_errors |
train_r2 |
test_r2 |
|
|---|---|---|---|---|---|---|
mean |
0.04722 |
0.0906 |
0.0307 |
0.06218 |
0.978 |
0.91972 |
min |
0.0386 |
0.0884 |
0.0244 |
0.0602 |
0.9729 |
0.912 |
max |
0.0529 |
0.093 |
0.0348 |
0.0633 |
0.9855 |
0.9265 |
std |
0.00474148 |
0.00195038 |
0.00354514 |
0.00117881 |
0.00417852 |
0.00460191 |
MODNet - MATMINER+LOBSTER
train_rmse |
test_rmse |
train_errors |
test_errors |
train_r2 |
test_r2 |
|
|---|---|---|---|---|---|---|
mean |
0.03602 |
0.08594 |
0.02336 |
0.05874 |
0.98732 |
0.92784 |
min |
0.0348 |
0.0823 |
0.0223 |
0.0568 |
0.9859 |
0.9231 |
max |
0.0378 |
0.091 |
0.0249 |
0.0609 |
0.9882 |
0.9324 |
std |
0.00101863 |
0.00283591 |
0.00095205 |
0.0014207 |
0.000767854 |
0.00374785 |
Corrected resampled t-test on 10-fold CV#
Summary
t_stat |
p_value |
significance_stars |
d_av |
rel_improvement |
percent_folds_improved |
|
|---|---|---|---|---|---|---|
RF |
2.50417 |
0.0168154 |
* |
0.356097 |
1.47348 |
90 |
MODNet |
1.65115 |
0.0665515 |
0.551488 |
3.04386 |
80 |
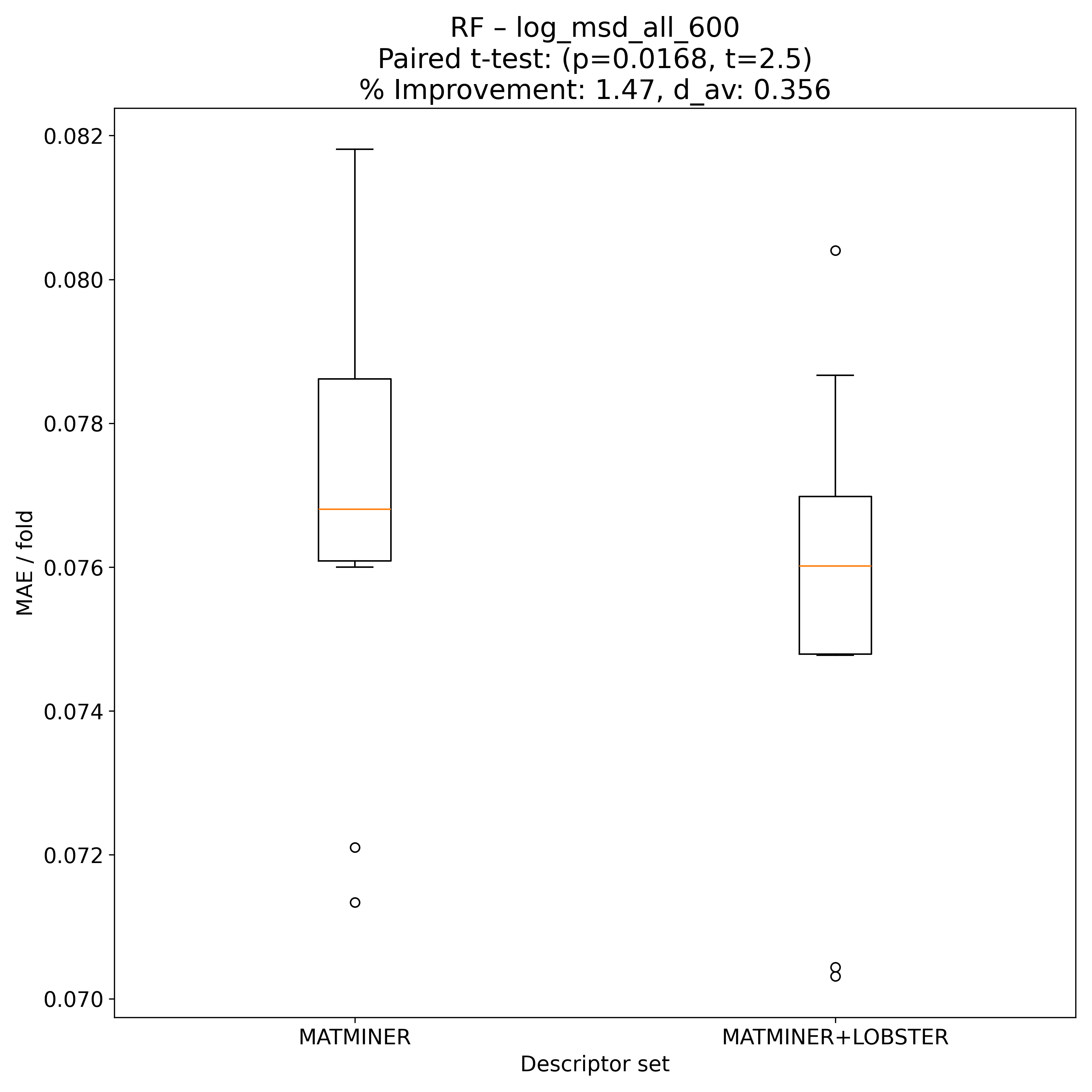
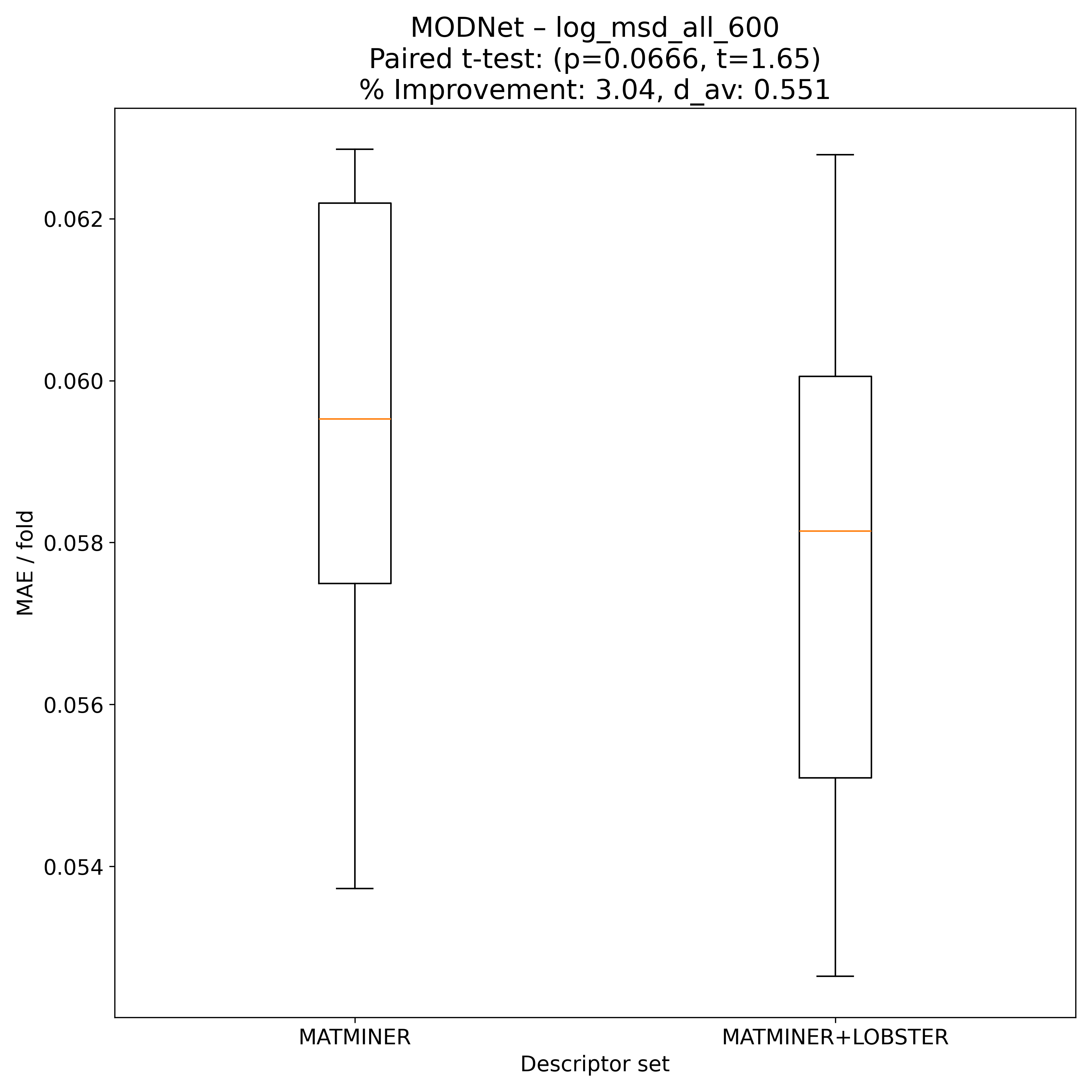
Model Explainer#
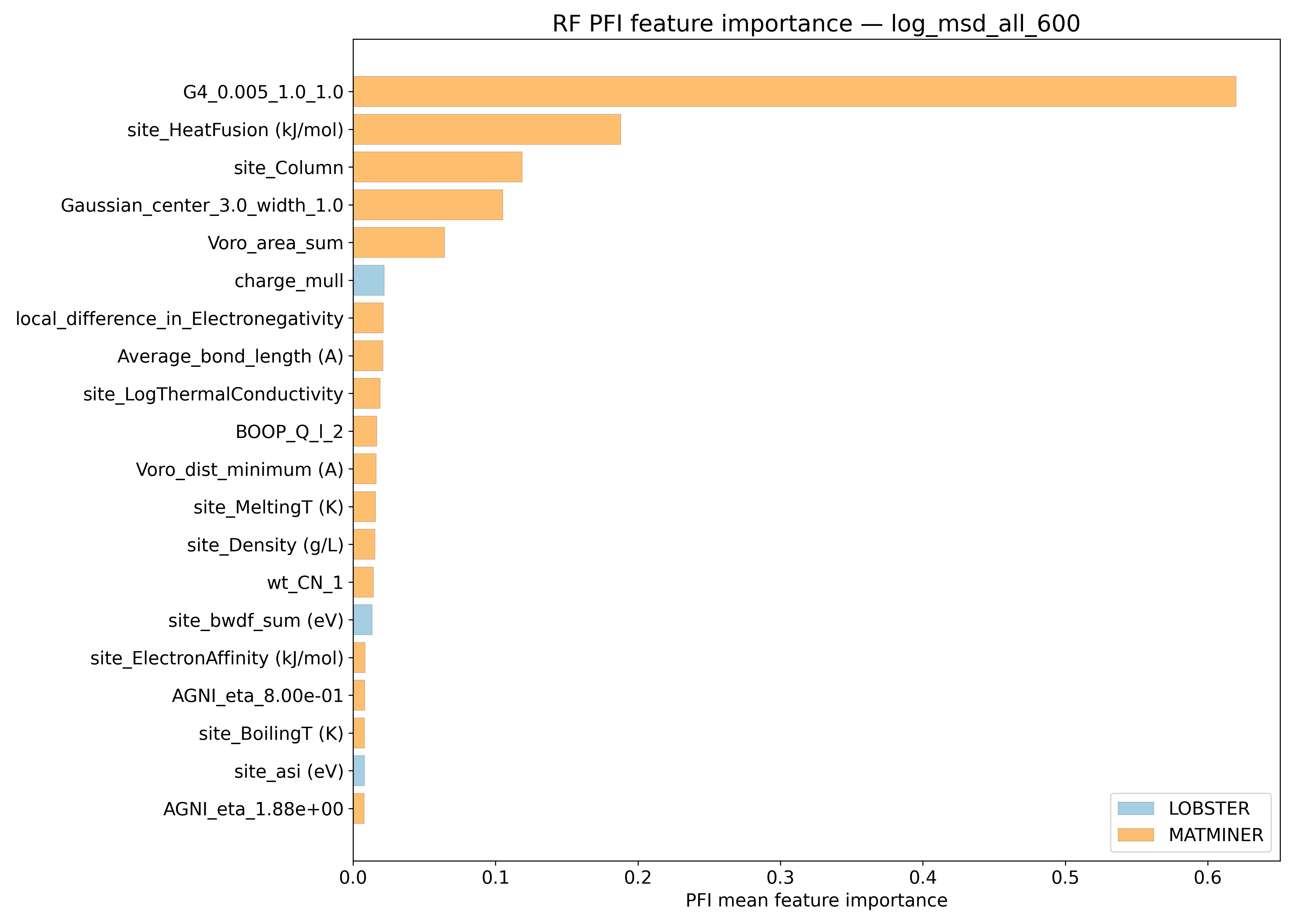
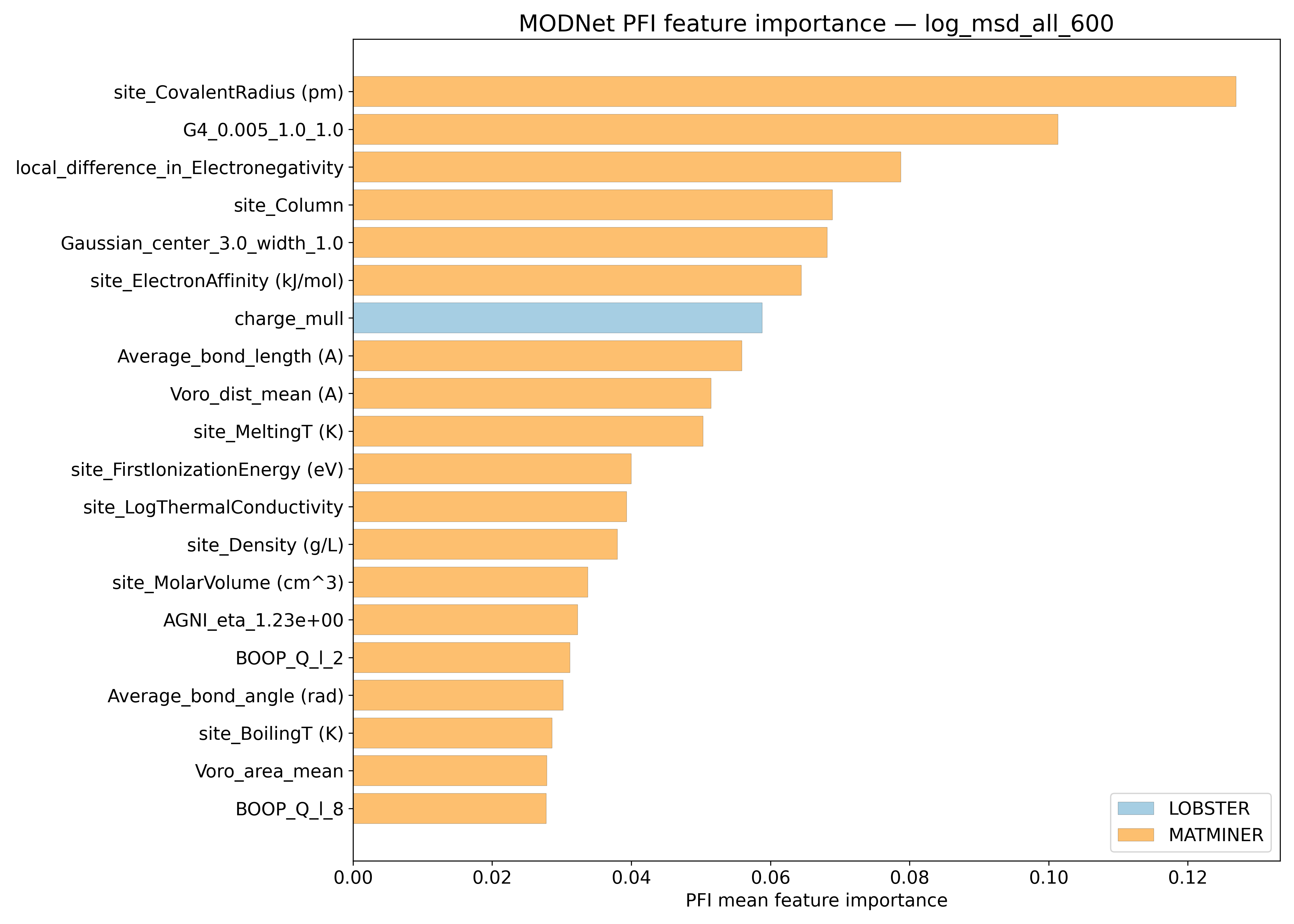
PFI#
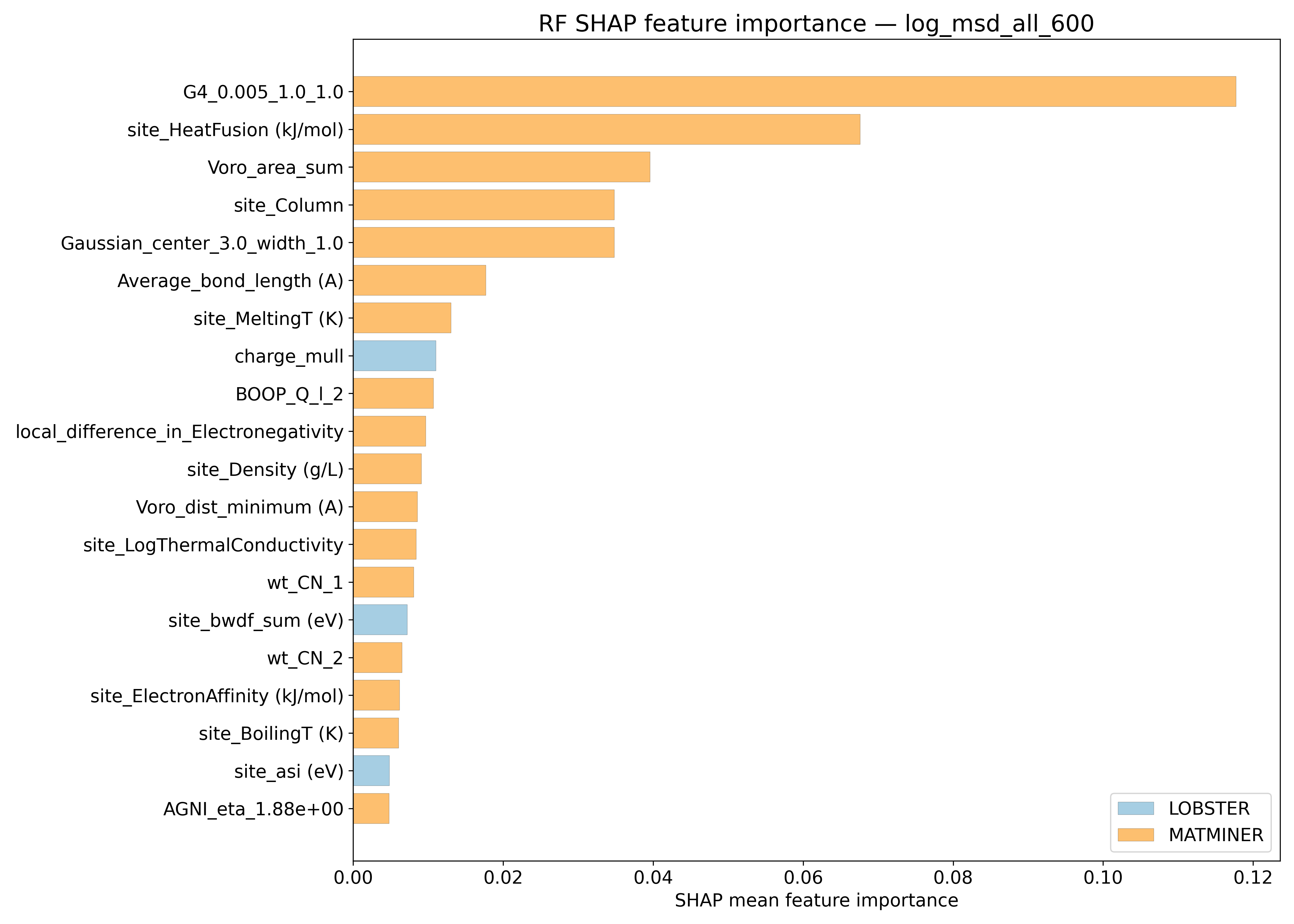
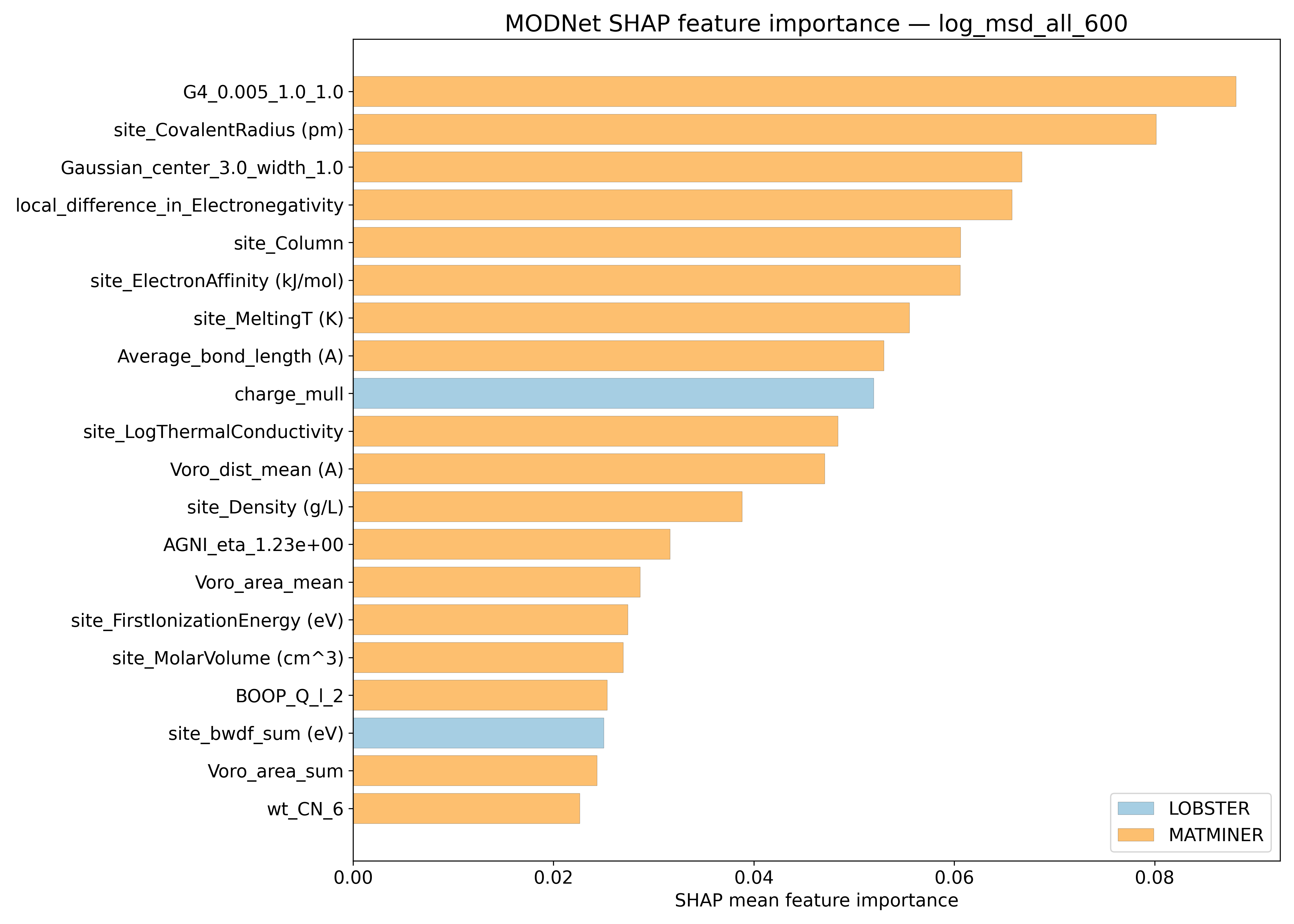
SHAP#
SISSO Models#
Rung 1#
1D descriptor#
2D descriptor#
Rung 2#
1D descriptor#
2D descriptor#
Misc#
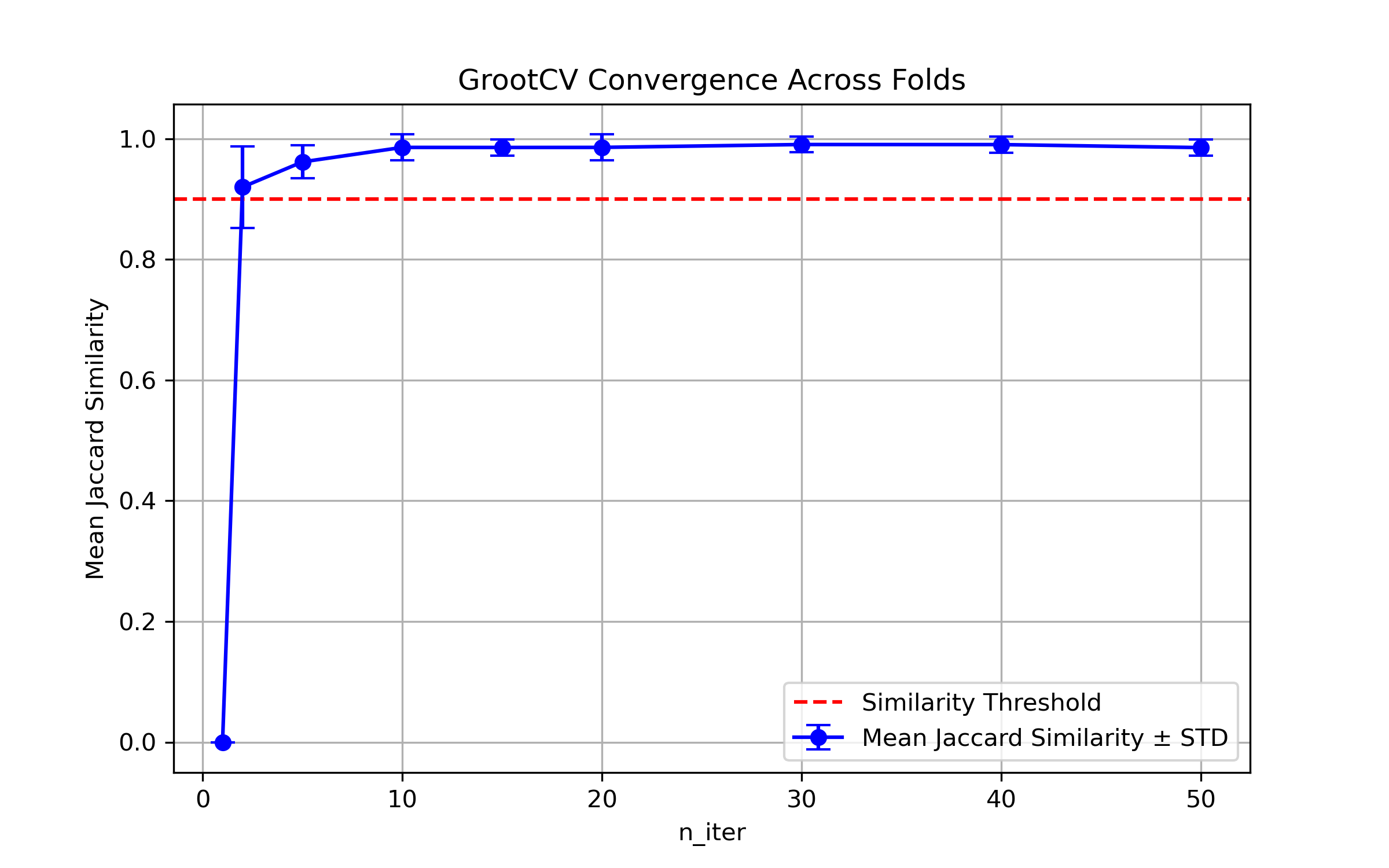
ARFS n-iter convergence checks#
MAE/ fold from 10-fold CV#
Alternative visual summary of input data for t-test