Log10 (All unique sites mean squared displacements @ 300K - Ų) - log_msd_all_300#
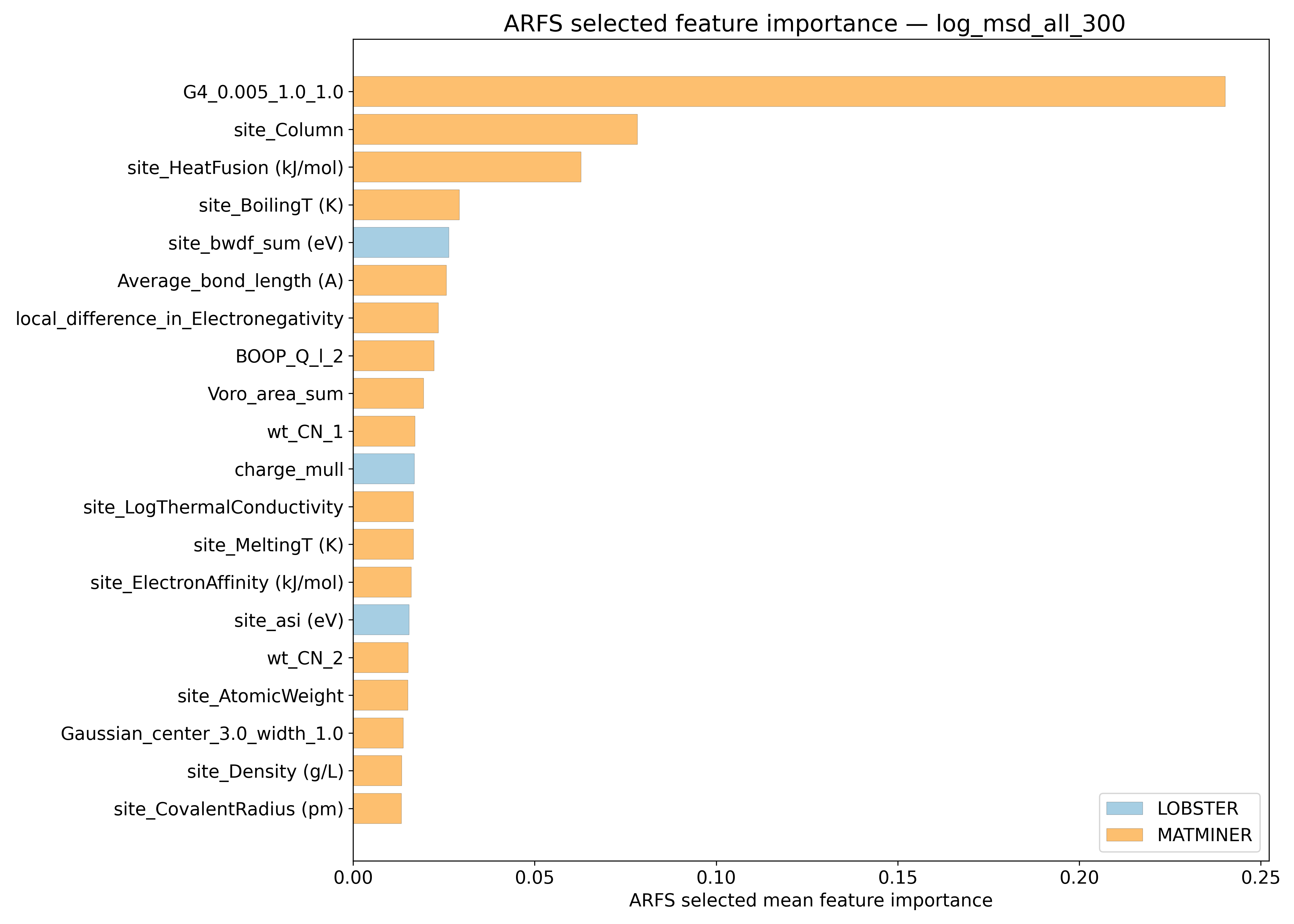
ARFS Top features#
ARFS selected descriptors#
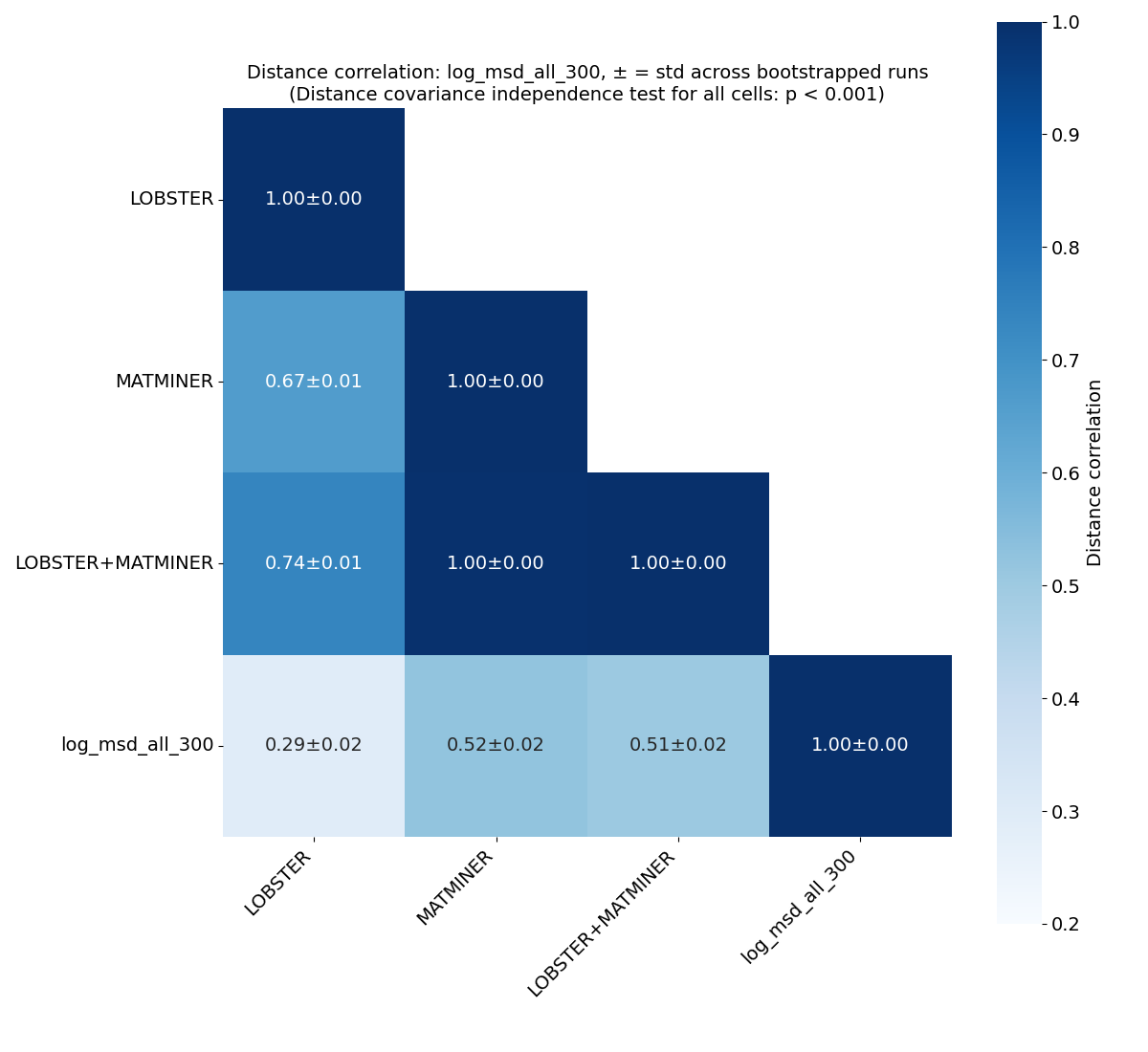
Correlation analysis#
Distance correlation#
Dependency graphs#
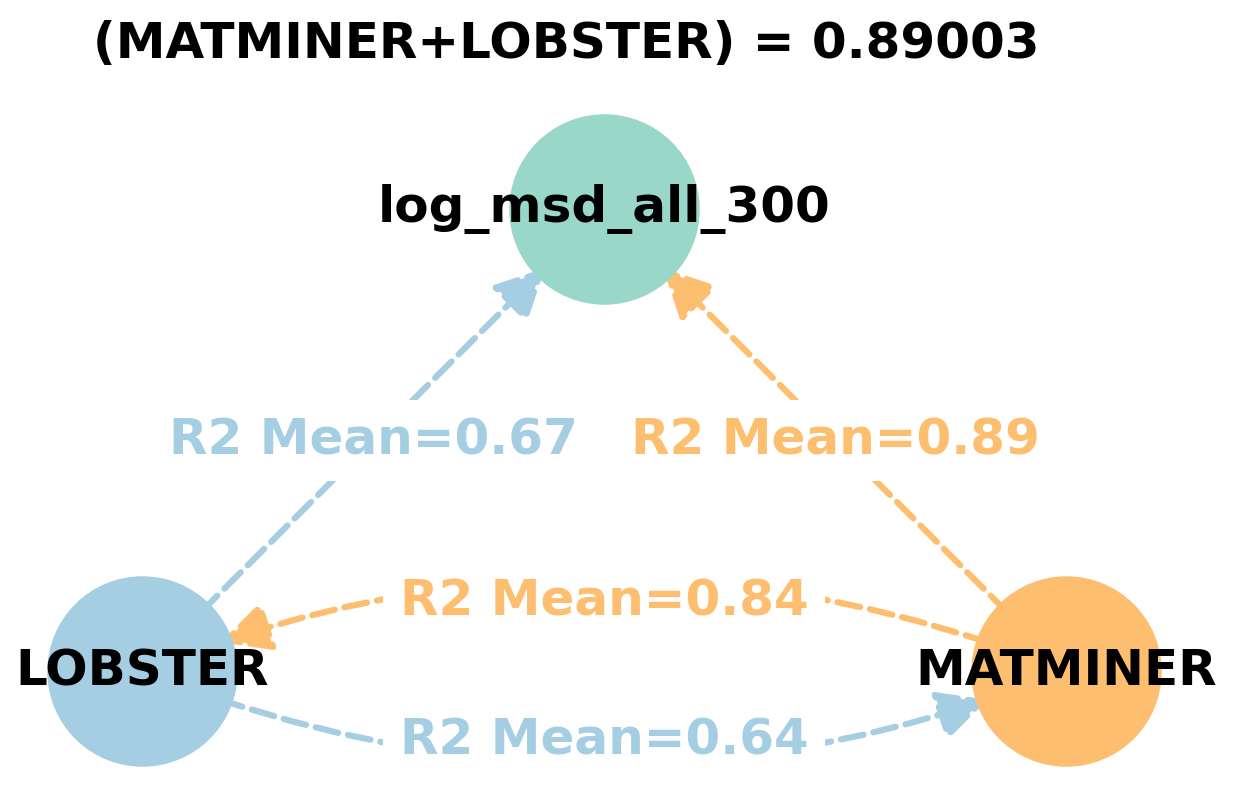
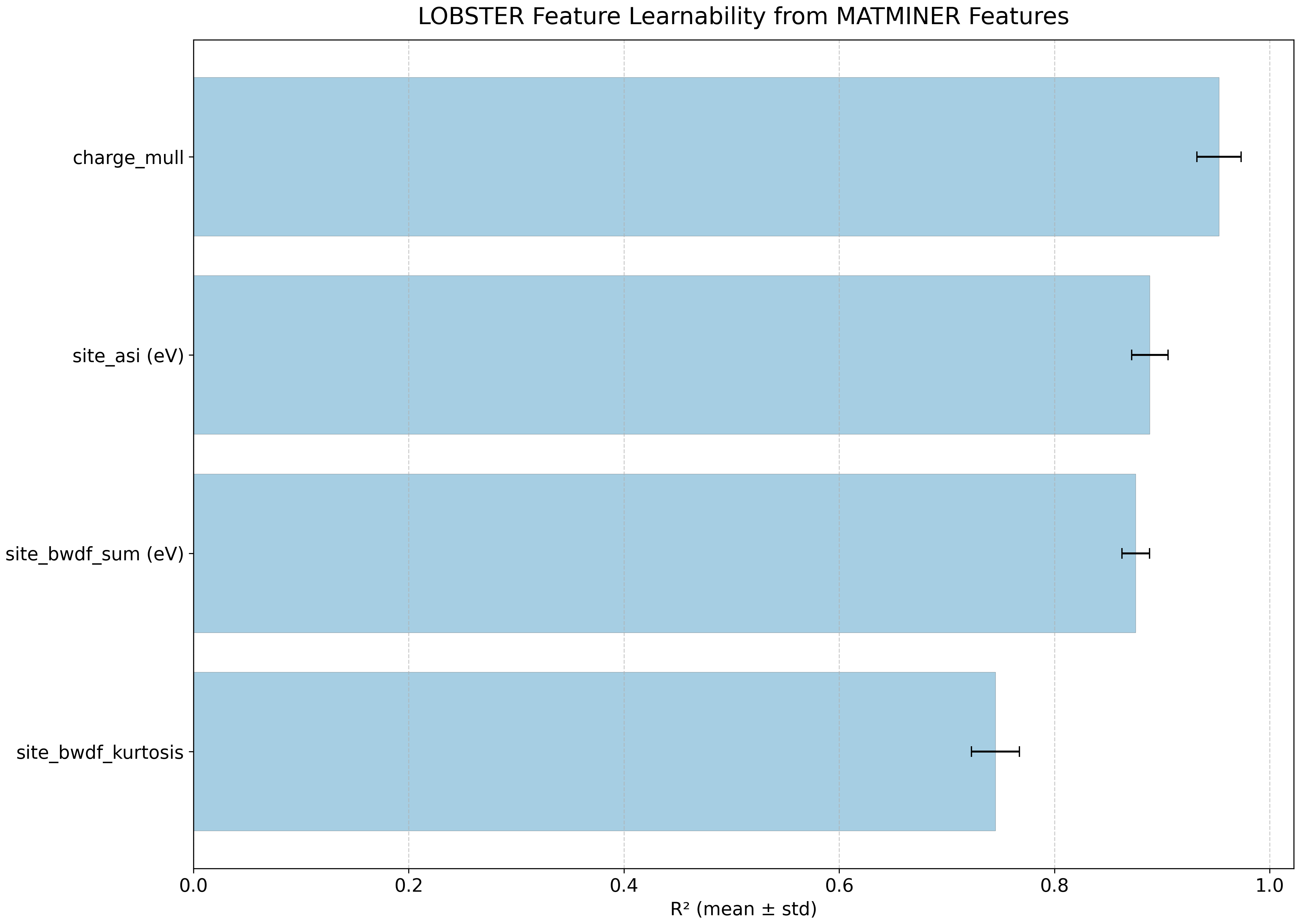
Feature learnability#
Model performance#
5-Fold CV Metrics overview#
RF - MATMINER
train_rmse |
test_rmse |
train_errors |
test_errors |
train_r2 |
test_r2 |
|
|---|---|---|---|---|---|---|
mean |
0.03926 |
0.10432 |
0.02832 |
0.07562 |
0.9841 |
0.8875 |
min |
0.0388 |
0.0999 |
0.0281 |
0.0748 |
0.9837 |
0.8737 |
max |
0.0396 |
0.1094 |
0.0286 |
0.0769 |
0.9846 |
0.9006 |
std |
0.00026533 |
0.00393517 |
0.000203961 |
0.000724983 |
0.000289828 |
0.00901887 |
RF - MATMINER+LOBSTER
train_rmse |
test_rmse |
train_errors |
test_errors |
train_r2 |
test_r2 |
|
|---|---|---|---|---|---|---|
mean |
0.03856 |
0.10306 |
0.0278 |
0.0747 |
0.98468 |
0.8902 |
min |
0.038 |
0.098 |
0.0275 |
0.0728 |
0.9844 |
0.8758 |
max |
0.0391 |
0.1089 |
0.0282 |
0.077 |
0.9852 |
0.9015 |
std |
0.000361109 |
0.00464052 |
0.000289828 |
0.00146697 |
0.000318748 |
0.00958374 |
MODNet - MATMINER
train_rmse |
test_rmse |
train_errors |
test_errors |
train_r2 |
test_r2 |
|
|---|---|---|---|---|---|---|
mean |
0.0393 |
0.08646 |
0.0251 |
0.05858 |
0.98392 |
0.92268 |
min |
0.0358 |
0.0816 |
0.0231 |
0.0572 |
0.9773 |
0.9167 |
max |
0.0471 |
0.0891 |
0.0305 |
0.0603 |
0.9867 |
0.9336 |
std |
0.00408363 |
0.00294252 |
0.00275318 |
0.00114263 |
0.00346318 |
0.00616876 |
MODNet - MATMINER+LOBSTER
train_rmse |
test_rmse |
train_errors |
test_errors |
train_r2 |
test_r2 |
|
|---|---|---|---|---|---|---|
mean |
0.04068 |
0.08644 |
0.02634 |
0.0592 |
0.98262 |
0.92286 |
min |
0.0342 |
0.0823 |
0.0222 |
0.0557 |
0.9774 |
0.9172 |
max |
0.047 |
0.0901 |
0.0304 |
0.0632 |
0.988 |
0.9284 |
std |
0.00534318 |
0.00298101 |
0.00351659 |
0.00300267 |
0.00447946 |
0.00402423 |
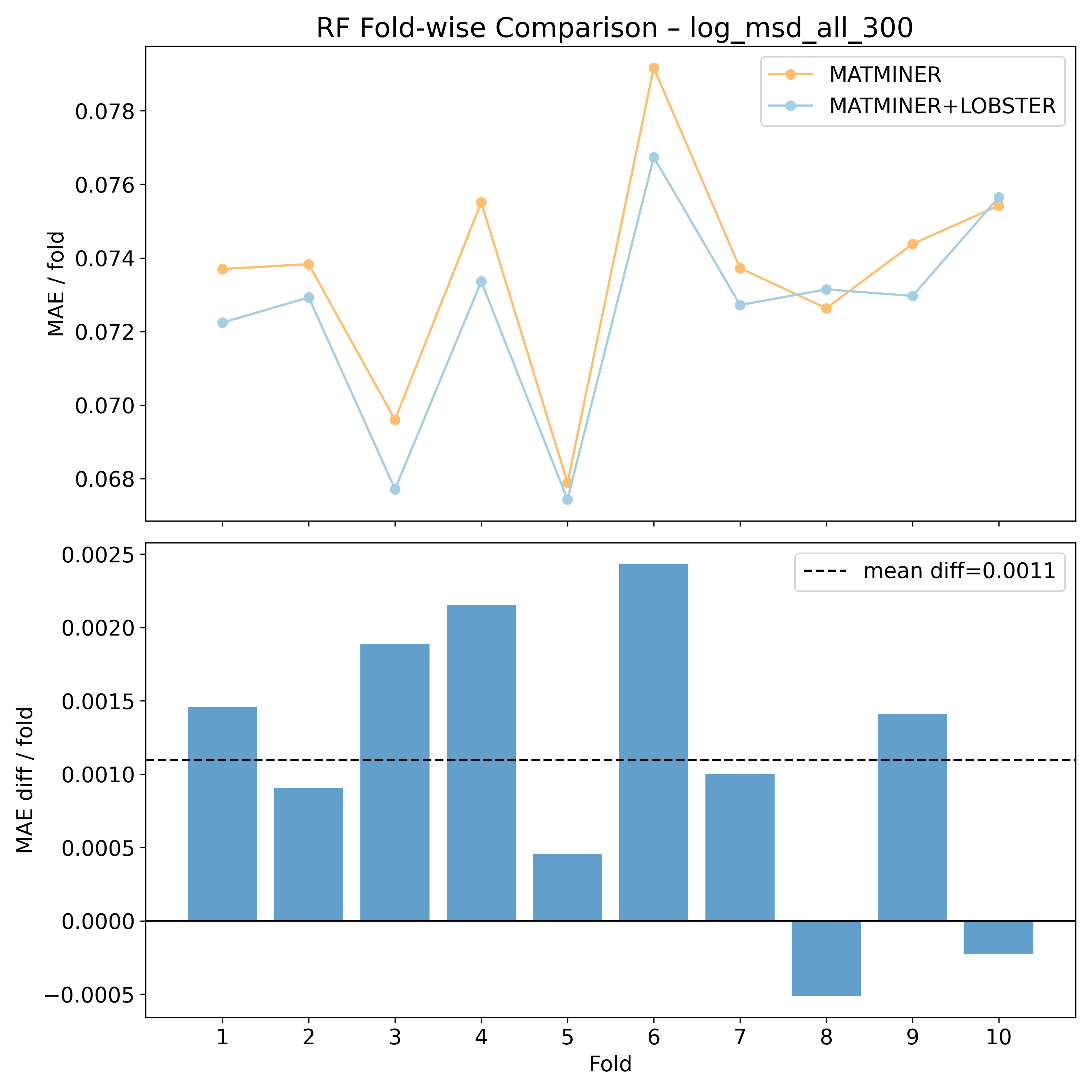
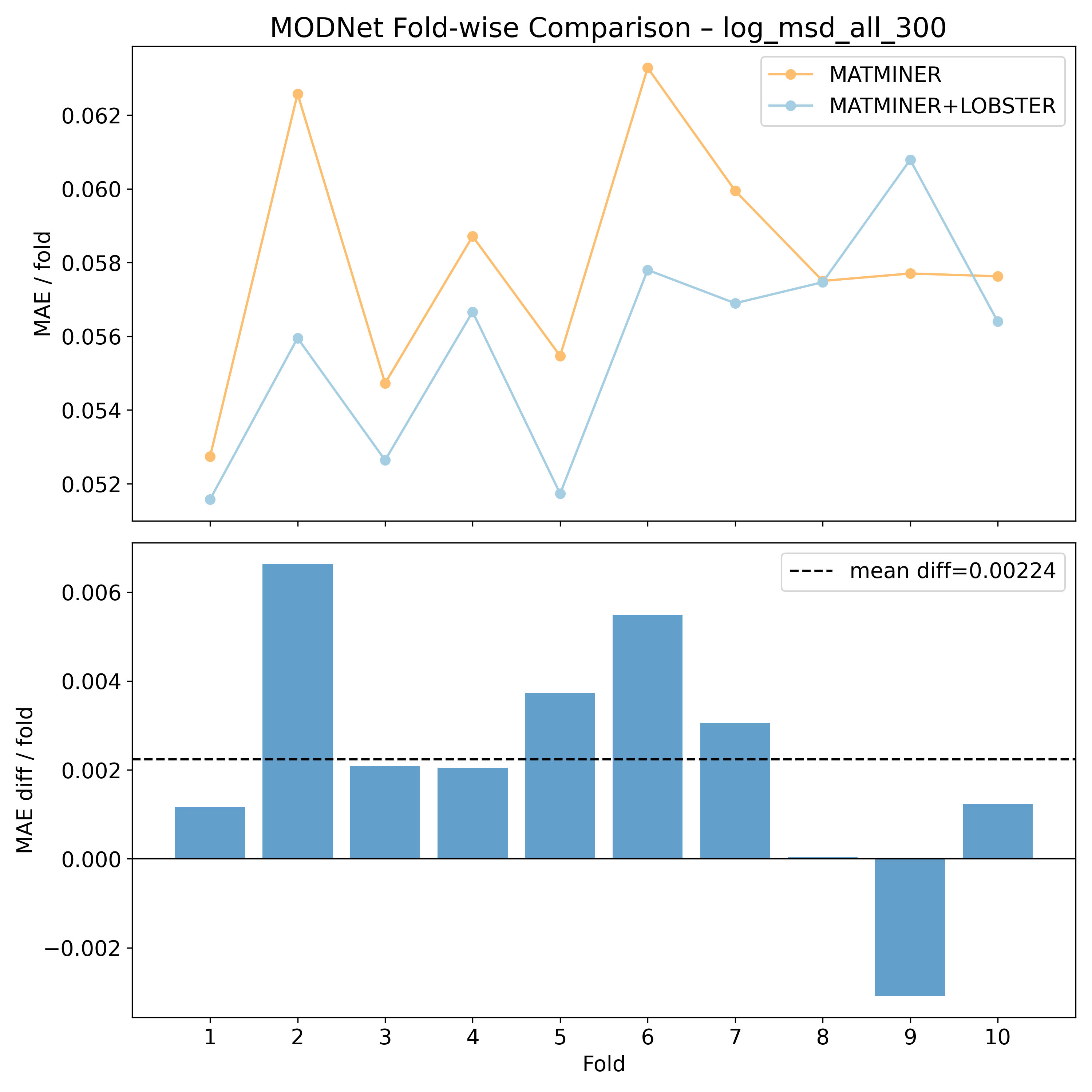
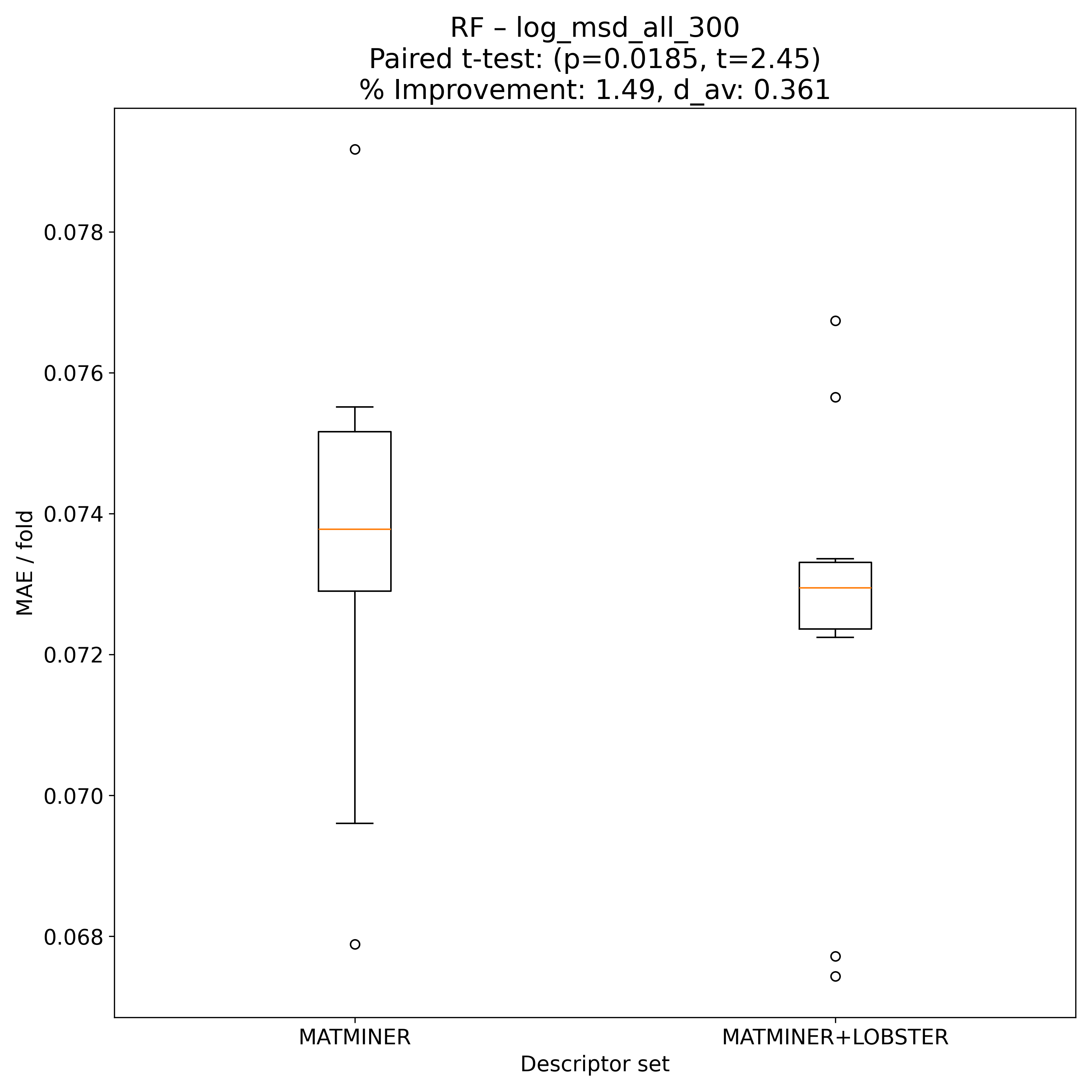
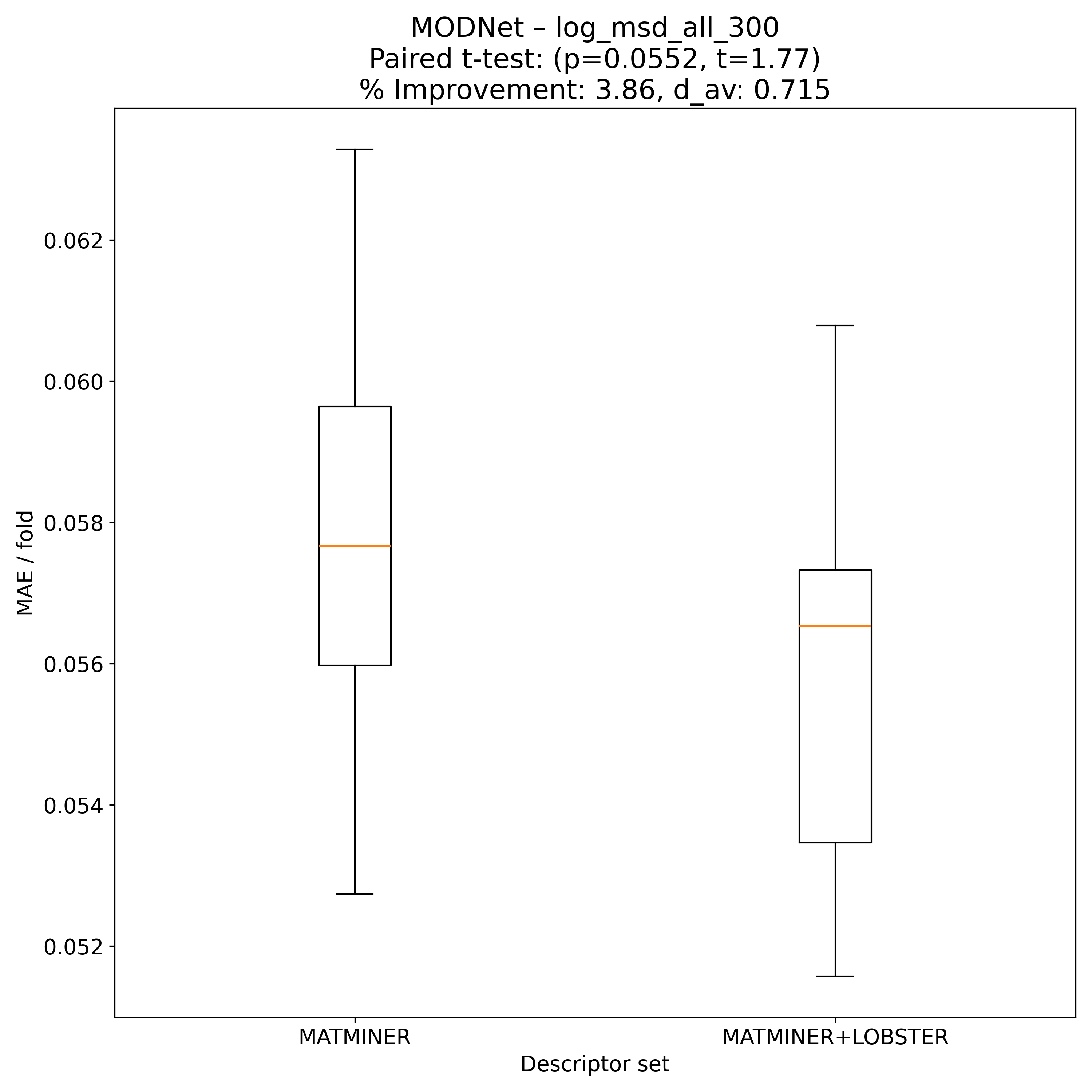
Corrected resampled t-test on 10-fold CV#
Summary
t_stat |
p_value |
significance_stars |
d_av |
rel_improvement |
percent_folds_improved |
|
|---|---|---|---|---|---|---|
RF |
2.44741 |
0.0184566 |
* |
0.360562 |
1.48878 |
80 |
MODNet |
1.77032 |
0.0552254 |
0.715069 |
3.85596 |
90 |
Model Explainer#
PFI#
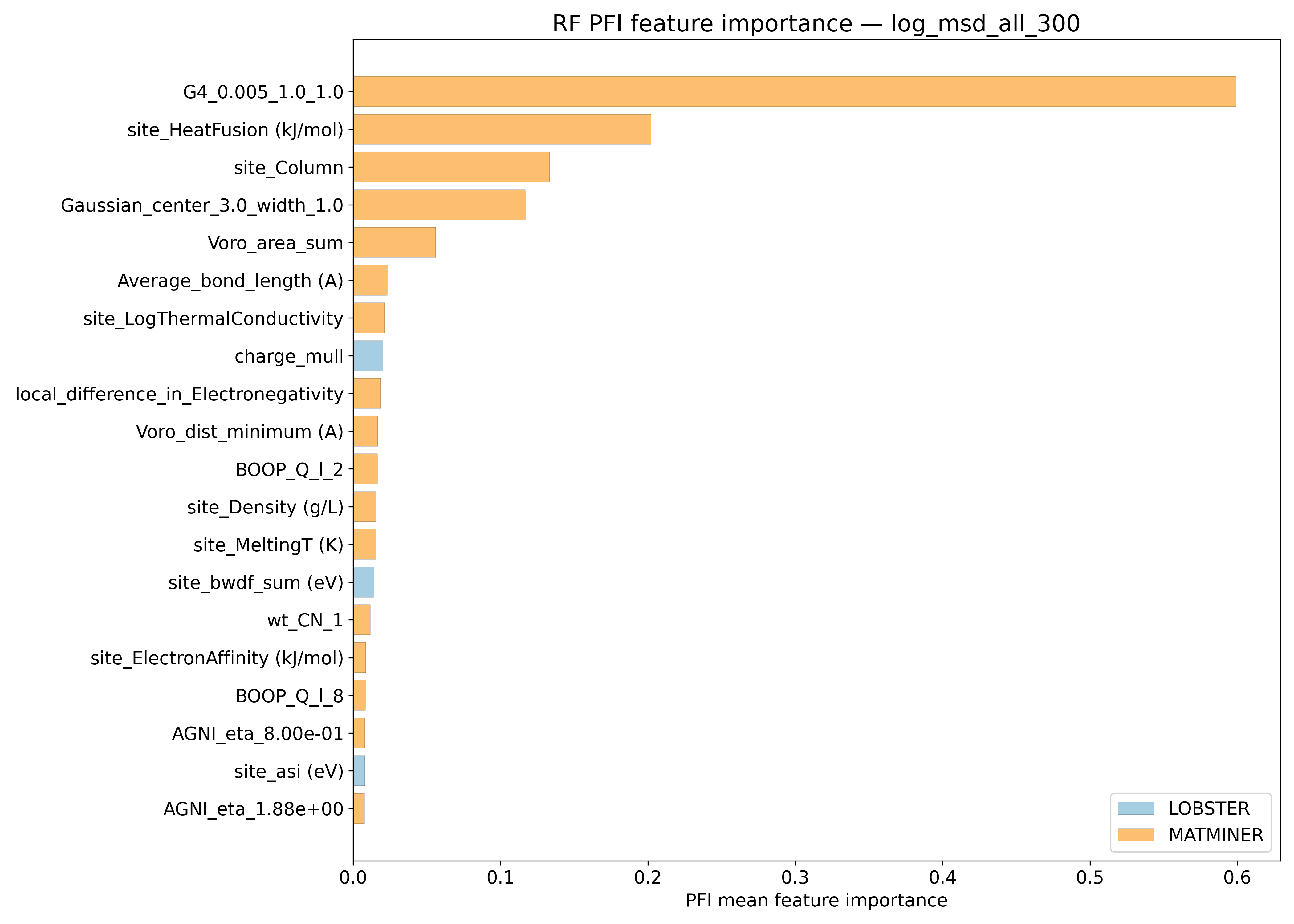
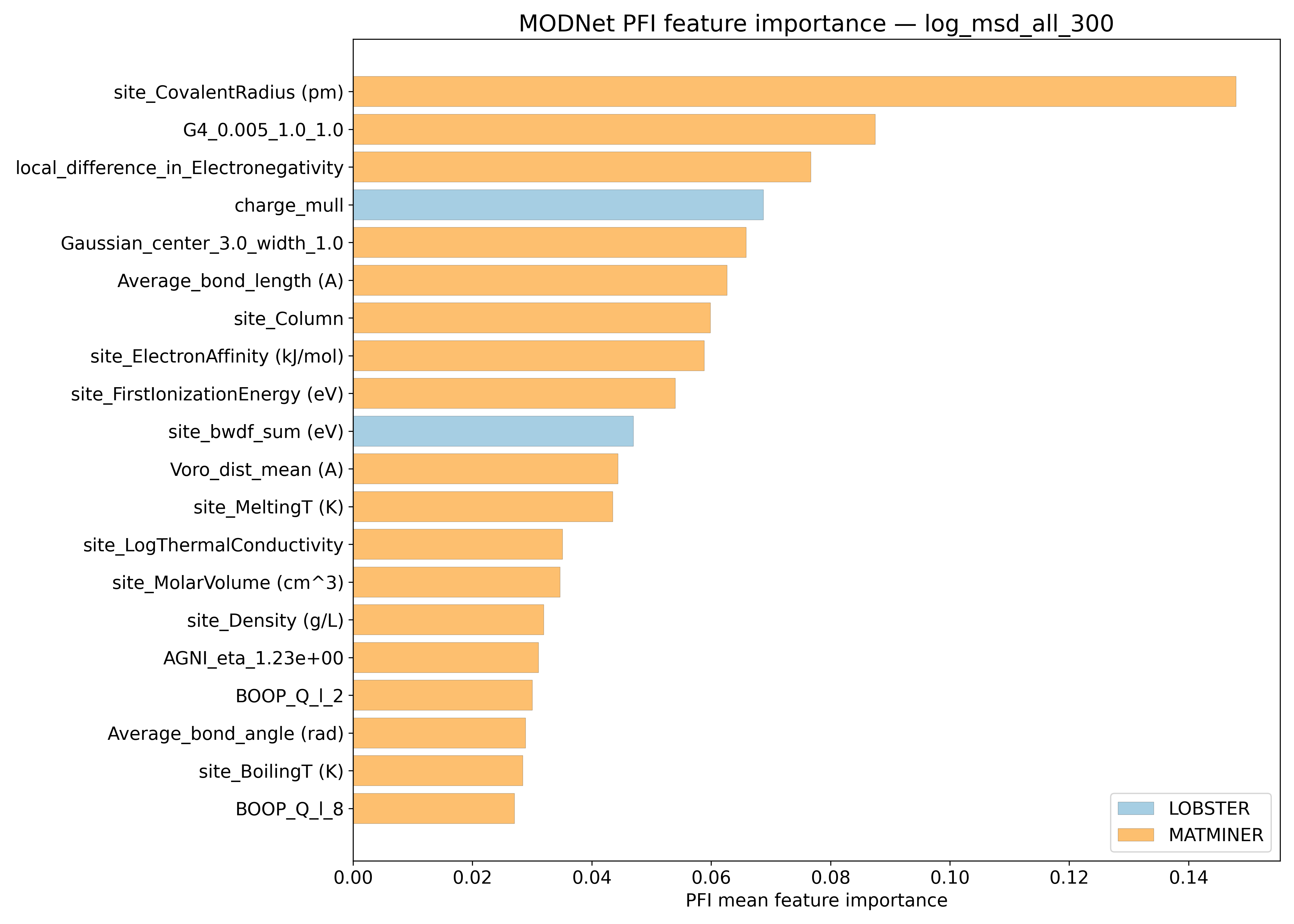
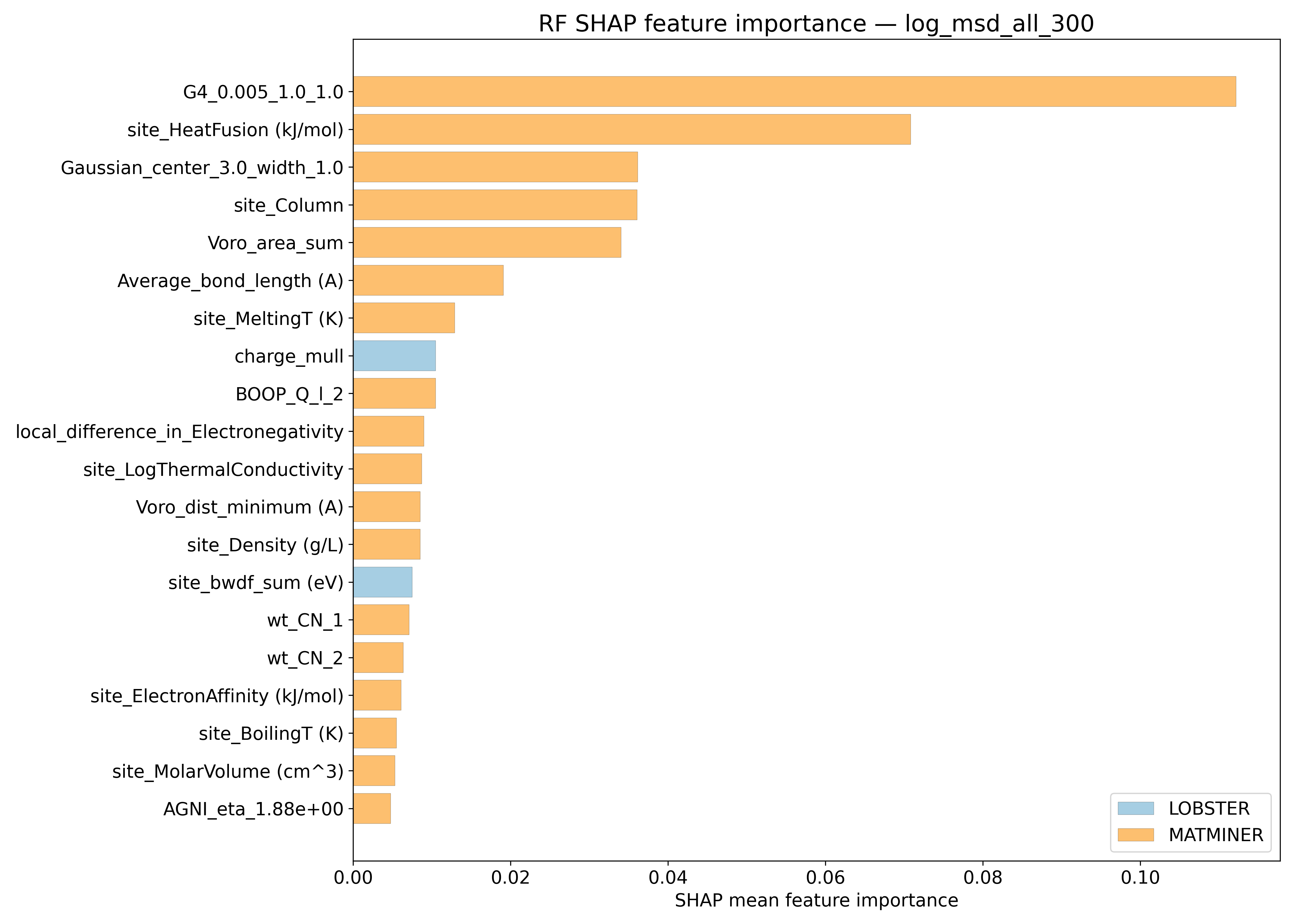
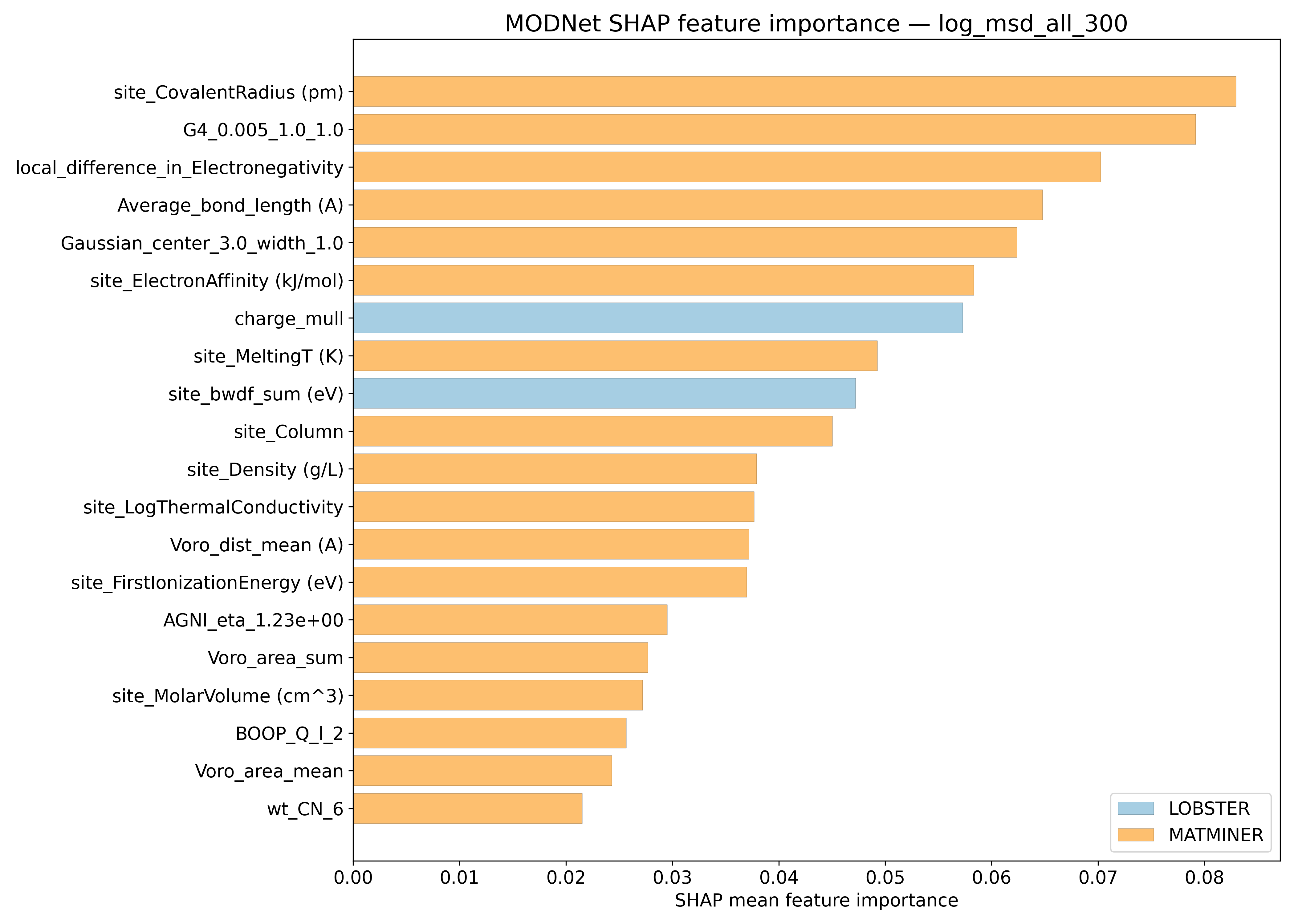
SHAP#
SISSO Models#
Rung 1#
1D descriptor#
2D descriptor#
Rung 2#
1D descriptor#
2D descriptor#
Misc#
ARFS n-iter convergence checks#
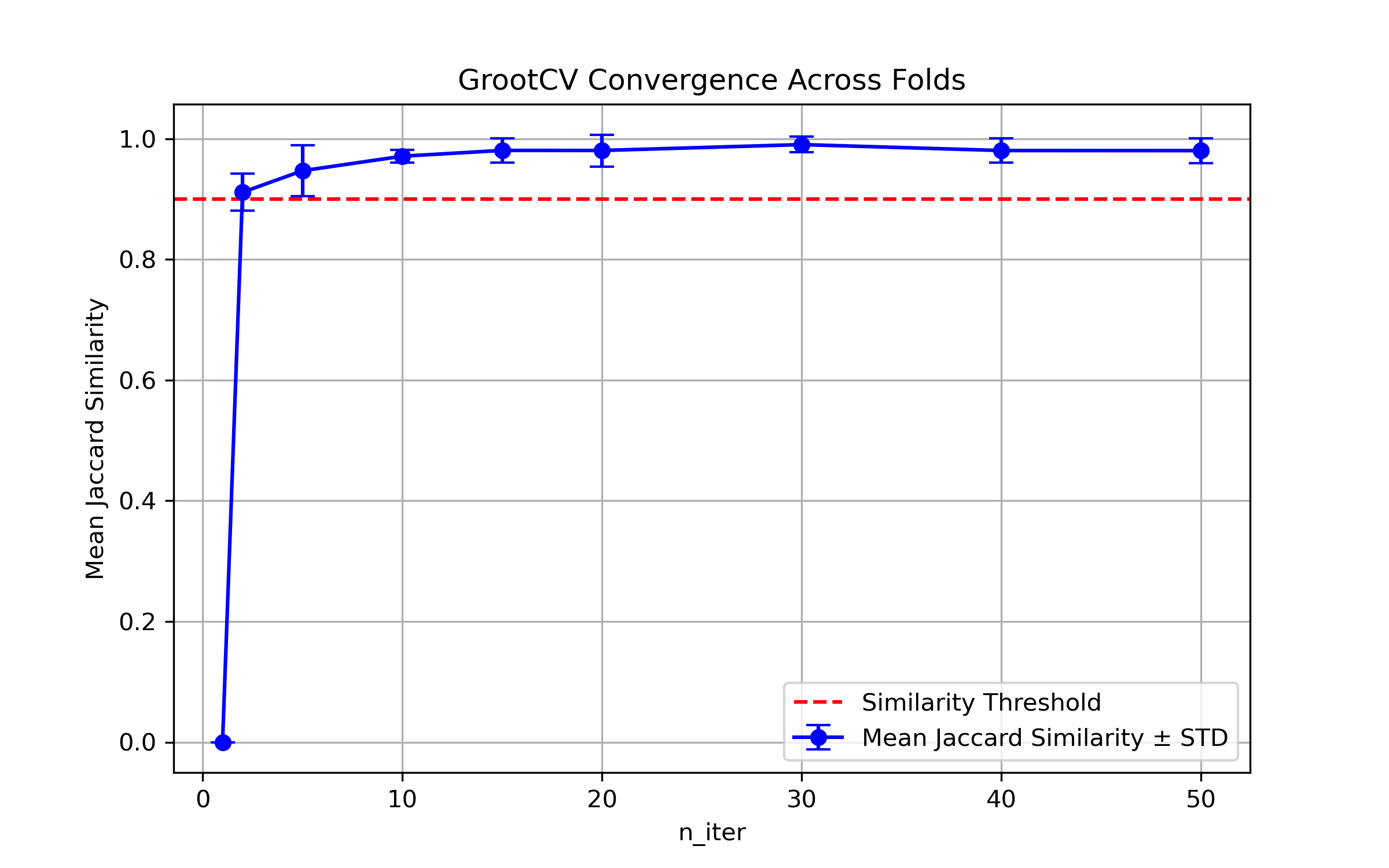
MAE/ fold from 10-fold CV#
Alternative visual summary of input data for t-test